Beta version
Introduction
Xwalk allows the user to validate and predict cross-link data by calculating Solvent Accessible Surface Distances (SASD) between amino acids. Please click on the various sections of the following two overview images to navigate through this help page. The images show an examplary run of Xwalk on the rabit fructose-bisphosphate aldolase A (ALDOA_RABIT) in Validation Mode (green circle in the Running Mode figure). The Xwalk run demonstrates that only 8 out of 14 intra-protein cross-links can be validated on the proteins X-ray structure (3DFQ).

Home Page
Running Mode
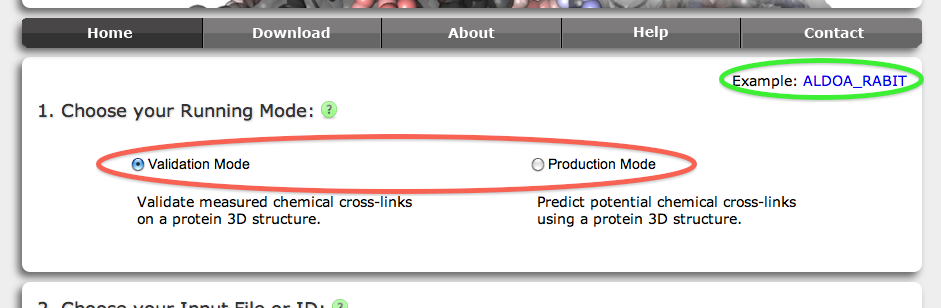
Xwalk can be executed in Validation Mode and Production Mode (red circle).
In Validation Mode the user can provide Xwalk a list of residue number pairs that the experimentalist found to be cross-linked or to be of interest. Given a maximum distance, Xwalk will calculate their SASD and report the SASD together with additional information for those residue number pairs that have their distance smaller than maximum distance.
In Production Mode Xwalk will report a list of in silico predicted virtual cross-links (vXL) that have some probability of being observed in a cross-linking experiment. The probability is given in analogy to Bernoulli trials solely by the amino acid pairs having a SASD distance smaller than maximum distance. Given a maximum distance, Xwalk will compute all intra-protein, inter-protein or both types of vXL.
Input
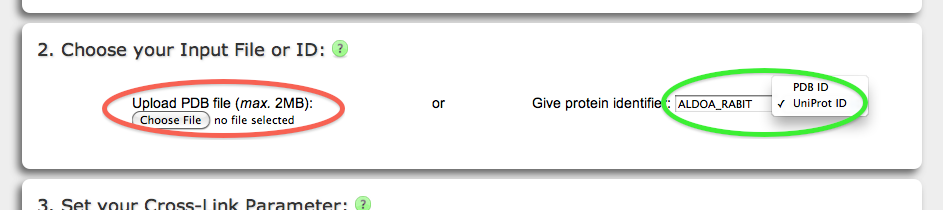
Xwalk accepts a protein either in form of a structure file in PDB format (red circle) or identifiers of the UniProt and PDB databases (green circle). Should the user provide a UniProt identifier in Validation Mode, (s)he will have also the option to specify the amino acid numbers with respect to the UniProt sequence. Otherwise, Xwalk will require the amino acid numbers as given in the PDB file of the protein.
Cross-link Parameter
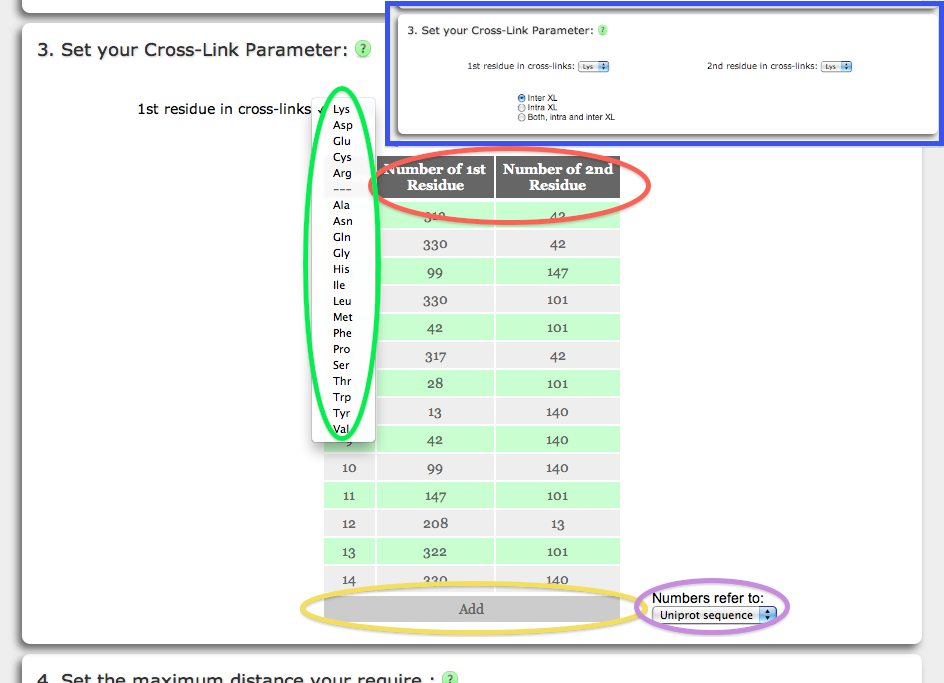
Xwalk can calculate vXL between any two amino acid pairs. One of the 20 standard amino acid types are available from the drop-down menus (green circle). The Validation mode is characterized by an additional limitation to amino acids with numbers given in the residue number pair table (red circle). The amino acid numbers must correspond to the numbering scheme in the PDB file, unless the user provides a UniProt ID, in which case (s)he can also add UniProt sequence based amino acid numbers (purple circle).
By default the table provides space for 10 residue number pairs, which however can be extended by clicking the Add field (yellow circle). The Production mode has no limitations on specific amino acids in the protein, but restricts the choice to the PDB chain identifier of the amino acids to determine the type of the vXL (blue bordered inset). Prior to the SASD calculation, all amino acid side chain will be removed, the solvent radius will be increased to 2.0 Å to compensate for the side chain removal in the SAS calculation. Subsequently, the distance will be calculated between beta-carbon atom of the chosen amino acids (alpha-carbons in the case of glycine amino acids).
Distance
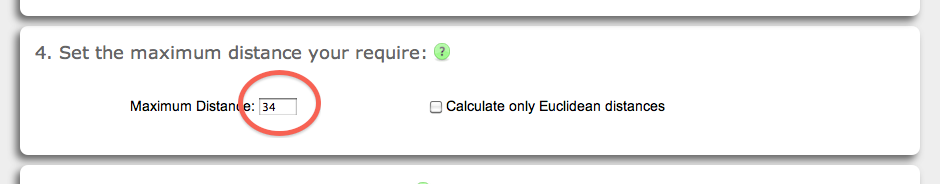
The SASD calculation is based on finding the shortest path between two points in a grid (see Implementation). Setting a maximum distance to the vXL calculation is nessaccary as the maximum distance defines the size of the grid. As larger the grid as longer the calculations last. Currently, the maximum supported distance is 60 Å for SASD calculations and 999 Å for Euclidean distance calculations. The default value for the maximum distance is 34 Å (red circle), which is the common upper bound on the SASD distances for Lys-Lys cross-links using disuccinimidyl suberate. Please note, that the SASD calculation include a solvent accessibility check of the cross-linked amino acids, while the Euclidean distance measure does not. On the other hand, Euclidean distance calculations are faster than SASD calculations (see below).
Result Page
Visualisation
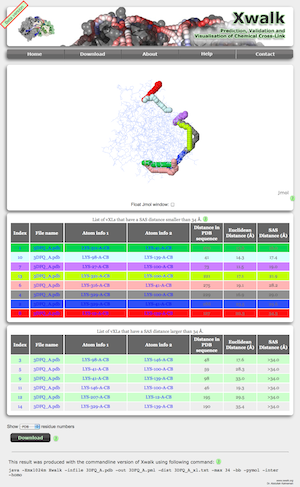
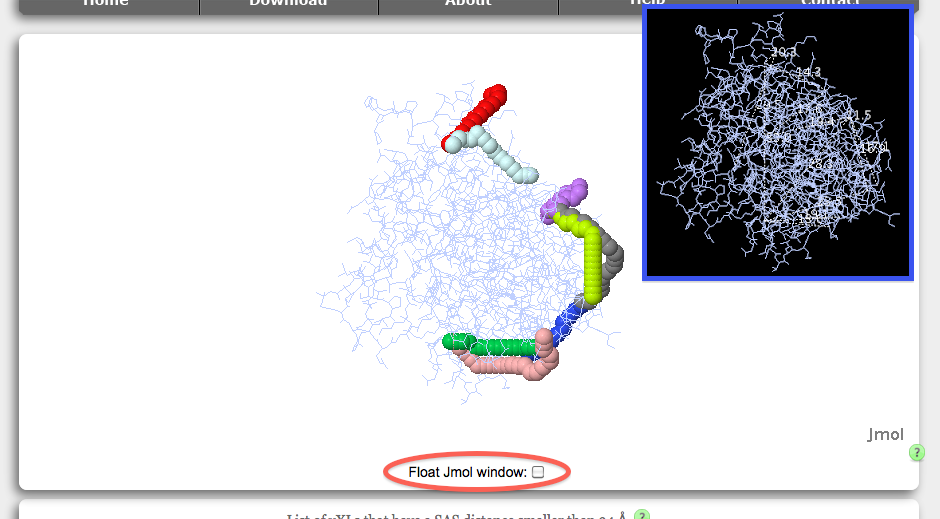
Once the distance calculations have been completed, Xwalk will map the vXL on the protein structure and visualise the mapping with Jmol: an open-source Java viewer for chemical structures in 3D. If your browser has problems initiating Jmol, please check your browsers Jmol compatibility here or refer to the system requirements of xwalk.org. Linux users might need to keep the floating function of the Jmol window switched off and reload the result page if the Jmol viewer crashes.
The protein structure is represented as a wireframe on a white background. A black background is used for Euclidean distance calculations to improve the readability of the distance information (blue bordered inset). Euclidean distances are displayed as white coloured dashed lines between the cross-linked amino acids. The shortest path from the SASD calculation are represented by a chain of pseudospheres, where each vXL path has its own color code. Translations, rotations and zoom around the protein are achieved via the computer mouse (see here for more details). Enabling the Float Jmol window check box (red circle), will keep the Jmol window fixed at the top border of the web page while scrolling down. Linux users might experience crashes of the Jmol applet when enabling the float functionality, in which case we recommend to reload the result page and keep the float option disabled.
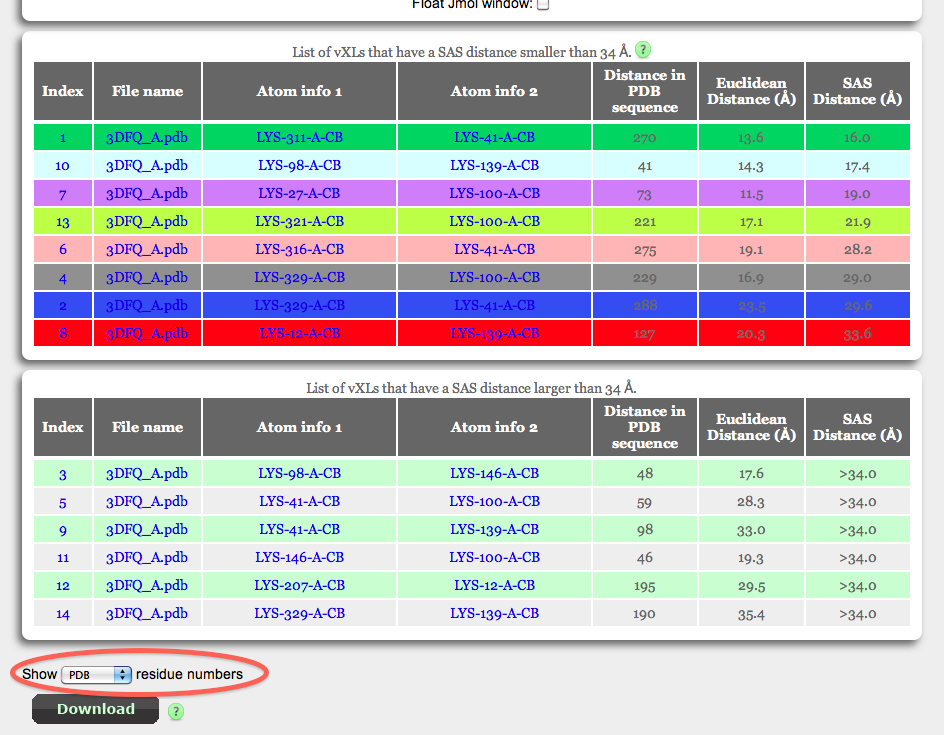
List of vXL
In addition to the visualisation of the vXL, Xwalk will also present a table with some useful information on each vXL (top table with multicoloured rows). The table is sorted according to SASD in ascending order, but can individually be sorted by clicking on one of the column headers. Amino acid pairs whos distance is beyond the maximum distance threshold are listed in a second table (bottom table with green and grey rows). The information in both tables includes:
- Index: In Production Mode, the index position is simply an iterator through all computed vXL. In Validation Mode, however the index number corresponds to the rank of the vXL in the residue number pair table.
- File Name: The PDB file name that was either uploaded by the user or retrieved from the PDB database based on the protein identifier (see Input).
- Atom info1/2: Details on the atoms that have been used for the vXL calculation. The information is divided by dashes and from left to right lists the amino acid name, PDB amino acid number, PDB chain Identifier and atom name. If the residue number pairs were specified as UniProt sequence number, the user will have also the option to display the UniProt amino acid number using the drop down menu at the bottom of the table.
- Distance in PDB sequence: This number is simply the difference of the positions of both amino acids in the vXL. This number, in particular for intra-protein cross-links is proportional to the amount of strutural information stored in the cross-link.
- Euclidean distance: The conventional distance measure between the beta-carbon atoms of the cross-linked amino acids. The distance calculation takes into account the flexibility of protein side chains in form of B-factors. This might have the effect that flexible residue pairs will conform to the maximum distance threshold even if they exceed it.
- SAS Distance: The new distance measure of Xwalk between the beta-carbon atoms of the cross-linked amino acids. The distance calculation takes into account the flexibility of protein side chains in form of B-factors. This might have the effect that flexible residue pairs will conform to the maximum distance threshold even if they exceed it.
The table contains hyperlinks to manipulate the view on the protein.
- Clicking on the Index number will show only the selected vXL on the proteins.
- Clicking on the Atom info1/2 link will show the selected amino acids in spacefill representation.
- Clicking on the File Name link will reset the view to the initial default view.
Download
All results, i.e. the PDB file, a PyMOL script for visualisation, the list of vXL and for Validation Mode runs, also the list of residue number pairs, can be downloaded by clicking on the download button

Shell Command
A commandline version of Xwalk written in JAVA is open source and available for free. The shell command at the bottom of the result page was placed to provide users the possibility to reproduce their results back on their local computer machines with the commandline version of Xwalk and where necessary using files from the download at the result page.
